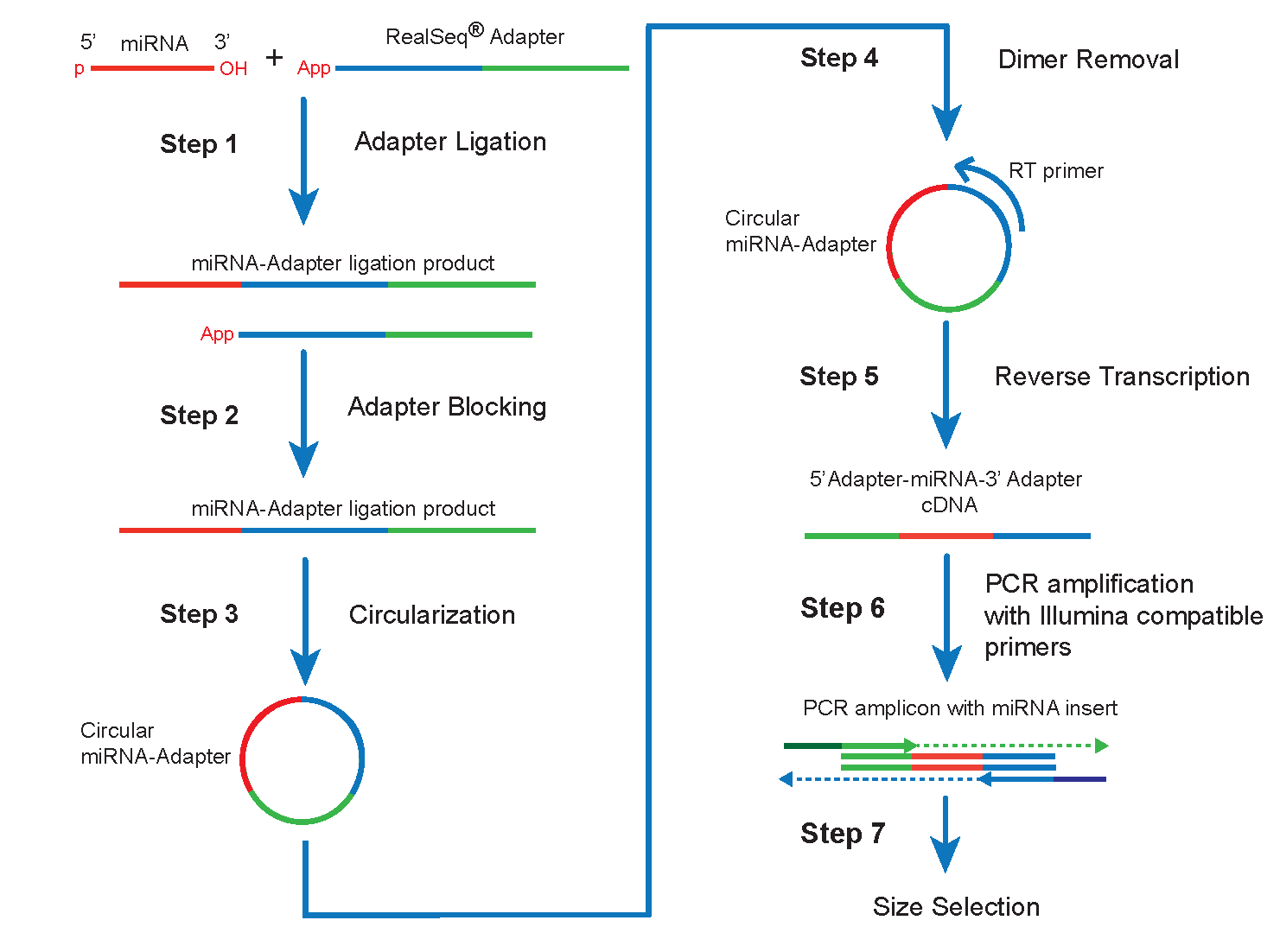
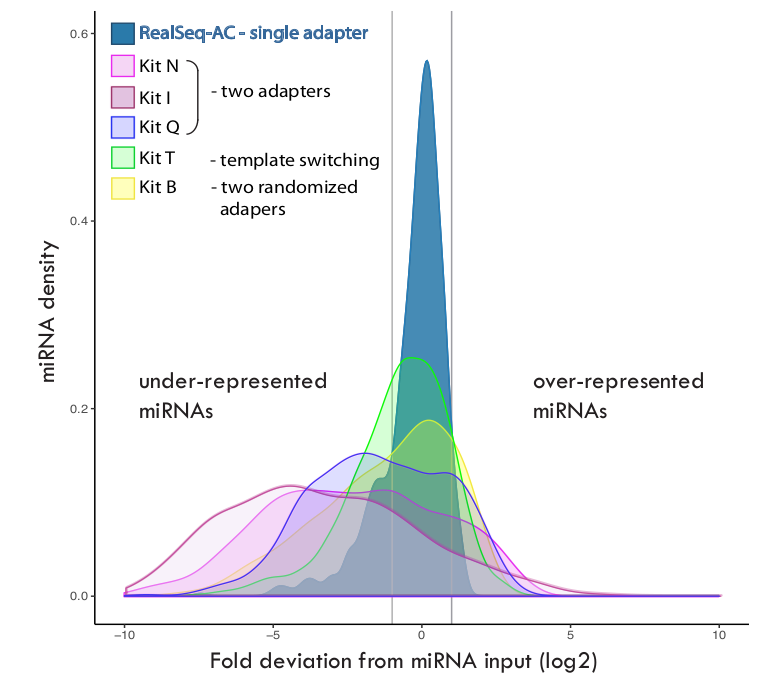
RiboMarker ® Platform
RiboMarkers are highly informative markers of disease and disease progression for conditions ranging from cancer to infectious diseases like funguses or viruses such as COVID. RiboMarkers can be used to determine not only whether a patient has a disease but also the state of the disease’s progression, for example, dissemination of an infection from the primary site throughout the body, markers of residual disease or possibly even of disease termination.
Our first RiboMarker diagnostic is for the fungal disease known as Valley Fever (VF). VF is a debilitating disease most often found in the Western states such as Arizona and CA and in Mexico and South America. When VF disseminates throughout the body patients require lifelong antifungal treatments that are costly and not very effective. The VF RiboMarker program allows better detection and analysis of dissemination and residual disease in these patients. VF and other “dimorphic” funguses diseases such as Histoplasmosis are dramatically increasing due to climate change.
Scientists at RealSeq are committed to expanding the RiboMarker diagnostic platform to address other fungal diseases as well as any other condition where the ability to accurately predict, identify, and track dissemination critically impacts patient outcomes.
Contact us (ribomarker@realseqbiosciences.com) if you are interested in discovering more with the RiboMarker Platform.